MONTREAL — A team of McGill University researchers has come up with a way to make playing video games an entirely productive pursuit.
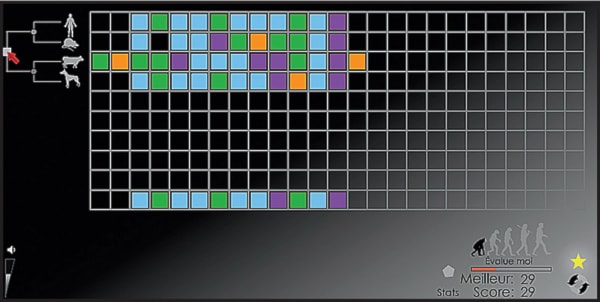
Computer scientist Jerome Waldispuhl, along with collaborator Mathieu Blanchette, developed a video game similar to the popular puzzle game Tetris — except there’s more to it than just stacking blocks.
The game, called Phylo, involves arranging sequences of coloured shapes that represent human DNA.
The game’s site says that when you play Phylo you are actually engaging in a wide framework to harness the power of many minds working to solve a common problem; multiple sequence alignments.
By analyzing the DNA sequences, scientists are able to gain new insight into the genetic basis of diseases like Alzheimer’s, diabetes and cancer. By studying these links biologists may infer shared evolutionary origins, or illustrate mutation events.
The game has enjoyed some popularity, with more than 17,000 people registering online since its November 2010 launch.
“Originally, I think a lot of people came because they are interested in participating in something that contributes to science,” Waldispuhl said in an interview.
“But the goal is really to expand that to casual players. People don’t really need to know that it contributes to science to enjoy it.”
The results of the players’ efforts can be retrieved by scientists from a database on the game’s website. So far, researchers have received more than 350,000 solutions to alignment sequence problems.
The game has already improved understanding of the regulation of 521 genes involved in a variety of diseases, Waldispuhl said.
Waldispuhl views the project as an attempt to combine the powers of the human brain with those of a computer.
There are some calculations that humans do more efficiently than any computer can, he said, such as recognizing and sorting visual patterns.
“As humans, we have evolved to handle visual information very efficiently,” he said.
By looking at the similarities and differences between these DNA sequences, scientists can get insight into genetically based diseases.
For example, one part of the game shows a human and a mouse, and the challenge is to align the nucleotides correctly in a gene connected with familial Alzheimer’s disease.
Once that is completed, the two sequences are compared with that of a dog and a new level of the game starts.
The trick is aligning the nucleotides — their order can’t be changed, but figuring out where along the sequence each one should go is a challenge, especially when one considers less-closely related species.
That kind of intuitive pattern recognition is not something computers are very good at.
This doesn’t mean humans can replace computers. In this instance, machines did a lot of the heavy lifting.
But the problem was the case of misaligned sequences, which the computers weren’t always able to spot.
While computers are best at handling large amounts of messy data, Waldispuhl said humans are able to sort through the coloured blocks representing DNA more quickly.
The genomes in the game were pre-aligned by computers, but parts remained slightly misaligned. As players sort out the puzzle in the game, they are also properly aligning the genomes.
Waldispuhl is hoping the game’s popularity continues to grow, creating a larger collection of data for genetic researchers.
With that in mind, Waldispuhl and his team recently created a version for tablets and smartphones.
“The idea is that it should be like Tetris, where people use it to take a break during the day,” he said.
“I would like them to play this instead of other games, because it’s like recycling the energy we are using.”
Here’s what one gamer noted on the web site for Genspace, a nonprofit organization dedicated to promoting education in molecular biology:
“I’ve played around with the DNA code responsible for idiopathic generalized epilepsy and already 160 other people attempted to solve the puzzle… 146 people failed.
“And there lies the problem of biology-turned games. You see, unlike regular puzzle games like Bejeweled or Tetris, not everything will fit together with perfect logical coherency.
“Granted, there are a few techniques you can use to treat this like any other game (for example, don’t waste your time moving around single blocks in the beginning stages. Crush them together into single group for maximum points in shortest amount of time), but the fact is not everything will fit together and it can be rather jarring for a beginner to figure out what he/she’s doing right, since there isn’t any satisfying feedback to a ‘correct’ sequence formation.
“It can’t be helped though. This is science, and no one knows the correct answers. They don’t exactly detect themselves and give you feedback. Maybe that’s the whole reason why you should play this game.
“After all, would you play Starcraft if the outcome were predetermined?”
To play the game or learn more about the research, visit http://phylo.cs.mcgill.ca/.